- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
FM1-43 Photoconversion and Electron Microscopy Analysis at the Drosophila Neuromuscular Junction
Published: Vol 7, Iss 17, Sep 5, 2017 DOI: 10.21769/BioProtoc.2523 Views: 9133
Reviewed by: Pengpeng LiAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Isolation of In Vitro Osteoblastic-Derived Matrix Vesicles by Ultracentrifugation and Cell-Free Mineralization Assay
Irshad A. Sheikh [...] Fayez K. Ghishan
Apr 5, 2025 1379 Views

Live Cell Imaging to Monitor Axonal Pruning in Drosophila Motor Neurons
Keyao Long [...] Menglong Rui
Jul 5, 2025 2038 Views

A Step-By-Step Protocol for Correlative Light and Electron Microscopy Imaging of Proteinaceous Deposits in Cultured Cells and Human Brain Tissues
Peizhou Jiang and Dennis W. Dickson
Aug 5, 2025 2343 Views
Abstract
We developed a protocol for photoconversion of endocytic marker FM1-43 followed by electron microscopy analysis of synaptic boutons at the Drosophila neuromuscular junction. This protocol allows detection of stained synaptic vesicle even when release rates are very low, such as during the spontaneous release mode. The preparations are loaded with the FM1-43 dye, pre-fixed, treated and illuminated to photoconvert the dye, and then processed for conventional electron microscopy. This procedure enables clear identification of stained synaptic vesicles at electron micrographs.
Keywords: Electron microscopyBackground
Neuronal transmitters are released via the fusion of synaptic vesicles with the neuronal plasma membrane. Vesicles can fuse spontaneously or in response to an action potential. Subsequently, vesicles become retrieved via endocytosis and recycled. Molecular mechanisms of synaptic vesicle recycling were investigated extensively with the tools of molecular biology, electrophysiology and microscopy (Slepnev and De Camilli, 2000; Sudhof, 2004; Rizzoli and Betz, 2005; Kavalali, 2006). Loading the endocytic marker FM1-43 coupled with the dye photoconversion followed by electron microscopy analysis is a powerful technique that allows the investigation and measurement of the recycling vesicle pools (Harata et al., 2001; Schikorski and Stevens, 2001; Rizzoli and Betz, 2004). Drosophila neuromuscular junction (NMJ) is an advantageous preparation with clearly defined synaptic boutons, which enables rapid generation of lines with mutated synaptic proteins and rigorous evaluation of vesicle recycling pools (Akbergenova and Bykhovskaia, 2009; Denker et al., 2009). A fundamental question in the field of synaptic transmission is whether the evoked and spontaneous transmission utilizes the same recycling pool. To address this question, the recycling pool utilized in the absence of stimulation needs to be measured. This is a challenging problem due to low rates of spontaneous release and recycling. We have developed a protocol for FM1-43 loading followed by the dye photoconversion and EM analysis, which enables rigorous evaluation of recycling pools utilized during spontaneous and evoked transmission at the Drosophila NMJ (Sabeva et al., 2017).
Materials and Reagents
- Sample preparation
- Gloves
- Long sleeve lab coat
- Sample bottle with snap cap, size 4 ml (Electron Microscopy Sciences, catalog number: 64250 )
- Falcon 35 mm culture plates (Corning, NY)
- Drosophila melanogaster fly stocks: Canton S (Bloomington Drosophila Stock Center, catalog ID: FBst1000081) and cpxSH1 (cpx-/-) (Huntwork and Littleton, 2007)
- Ca2+-free HL3 saline containing 75 μM advasep-7 (Biotium, catalog number: 70029 ) (see Note 1)
- FM1-43 (Thermo Fisher Scientific, InvitrogenTM, catalog number: T35356 ), 10 µM
- 100 mM NH4Cl (Sigma-Aldrich, catalog number: A9434 ) in HEPES
- 1.5 mg/ml DAB (Agilent Technologies, DAKO, OEM) in HEPES
Note: The compound is available liquid or tablets in kits. - 1% osmium tetroxide (OsO4) prepared in 90 mM cacodylate buffer using the 4% OsO4 stock solution (Electron Microscopy Sciences, catalog number: 19150 )
- Ascending acetone series 50, 70, and 90% prepared from 100%
- 2% uranyl acetate (Electron Microscopy Sciences, catalog number: 22400 ) prepared in water
- Acetone, 100% (Sigma-Aldrich, catalog number: 270725 ) dehydrated with molecular sieve dehydrate Fluka (Fluka Analysis, Sigma-Aldrich, catalog number: 270725 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S9888 )
- Potassium chloride (KCl) (Sigma-Aldrich, catalog number: P3911 )
- Magnesium chloride hexahydrate (MgCl2·6H2O) (Sigma-Aldrich, catalog number: M9272 )
- Calcium chloride dihydrate (CaCl2·2H2O) (Sigma-Aldrich, catalog number: C5080 )
- Sodium bicarbonate (NaHCO3) (Sigma-Aldrich, catalog number: S6014 )
- Trehalose (Sigma-Aldrich, catalog number: T9531 )
- Sucrose (Sigma-Aldrich, catalog number: S1888 )
- HEPES-Na salt (Sigma-Aldrich, catalog number: H7006 )
- HEPES (Sigma-Aldrich, catalog number: H3375 )
- Paraformaldehyde (Electron Microscopy Sciences, catalog number: 15710 )
- Glutaraldehyde (Electron Microscopy Sciences, catalog number: 16220 )
- Sodium cacodylate buffer (Electron Microscopy Sciences, catalog number: 11653 )
- HL3 Drosophila saline solution (see Recipes)
- Pre-fixative solution (see Recipes and Note 2)
- HEPES-buffered saline (see Recipes)
- Fixative solution (see Recipes and Note 2)
- Gloves
- Sample embedding
- ACLAR® 33C Embedding film (Electron Microscopy Sciences, catalog number: 50425-10 )
- Tri-Corn beakers: plastic, disposable (Electron Microscopy Sciences, catalog number: 60972 )
- Flat Bottom Embedding Capsules (Electron Microscopy Sciences, catalog number: 70021 )
- Epon (Embed-812) (Electron Microscopy Sciences, catalog number: 14900 )
- Nadic Methyl Anhydride (NMA) (Electron Microscopy Sciences, catalog number: 19000 )
- Dodecenyl Succinic Anhydride Specially Distilled (DDSA) (Electron Microscopy Sciences, catalog number: 13710 )
- 2,4,6-Tri(dimethylaminomethyl) phenol (DMP-30) (Electron Microscopy Sciences, catalog number: 13600 )
- ERL-4221 (Electron Microscopy Sciences, catalog number: 15004 )
- D.E.R. 736 Epoxy Resin (Used to simplify infiltration in combination with Embed 812) (Electron Microscopy Sciences, catalog number: 13000 )
- Nonenyl Succinic Anhydride Modified (NSA) (Electron Microscopy Sciences, catalog number: 19050 )
- Dimethylaminoethanol (DMAE) (Electron Microscopy Sciences, catalog number: 13300 )
- Embedding Mix A (see Recipes)
- Embedding Mix B (see Recipes)
- ACLAR® 33C Embedding film (Electron Microscopy Sciences, catalog number: 50425-10 )
Equipment
- Sample preparation
- Sylgard 184 (World Precision Instruments, Sarasota, FL)
- Fine tweezers (World Precision Instruments, models: #2 and #5 )
- Fine scissors (World Precision Instruments, model: 501233 )
- Insect Minutien Pins (0.1 mm) (Fine Science Tools, catalog number: 26002-10 )
- Dissecting stereoscopic zoom microscope (Nikon Stereozoom Microscope, Nikon Corporation)
- A.M.P.I. Master 8 Stimulator (A.M.P.I., model: Master-8 )
- Suction electrode filled with HL3
- Epifluorescence compound microscope with a filter cube with a long-pass emission filter customized for FM1-43 dye
- 60x water-immersion objective (ZEISS, Thornwood, NY)
- Mercury lamp
- Biowave (Ted Pella, Redding, CA)
- 480 ± 10 bandpass excision filter
- PELCO® R1 Single Speed Rotator in the 35° positions (Ted Pella, model: PELCO® R1 )
- Fume hood
- Sylgard 184 (World Precision Instruments, Sarasota, FL)
- Sample embedding
- Oven Binder (Tuttlingen, Germany)
- Oven Binder (Tuttlingen, Germany)
- Ultrathin Sections preparation and Image Capture
- Ultramicrotome for ultrathin sectioning (Leica, model: Leica EM UC6 )
- Formvar/carbon-coated 2 x 1 mm slot copper grids (Electron Microscopy Sciences, catalog number: FCF-2010-Cu )
- Ultra Diatome diamond sectioning knife, wet (Electron Microscopy Sciences, catalog number: 27-US )
- UltraTrim Dry Room Temperature Diatome knife, 4.0 mm (Electron Microscopy Sciences, catalog number: UTT-40-R )
- Eyelash with handle (Ted Pella, catalog number: 119 )
- Double edge stainless steel razor blades (Electron Microscopy Sciences, Hatfield, PA)
- Forceps for grids (Electron Microscopy Sciences, Hatfield, PA)
- JEOL 100 CX Electron Microscope equipped with Hamamatsu digital camera and AMT software
- Ultramicrotome for ultrathin sectioning (Leica, model: Leica EM UC6 )
Software
- ImageJ (National Institutes of Health) for image analysis
- Adobe Photoshop (Adobe Systems) software for image analysis
Procedure
- Dissection of Drosophila melanogaster larvae in HL3 solution
- Select third-instar larvae (L3) and place it in a Sylgard dish.
- Pin the L3 on the anterior (head/mouth apparatus) and the posterior part, and the dorsal side-up.
- Immerse the pinched larvae in 4-5 ml HL3 solution (Recipe 1) to the Sylgard dish.
- Section the dorsal side longitudinally and remove the internal organs (intestines, fat bodies and trachea).
- Stretch and pin the rest of the L3.
- Several muscles could be used. In this study experiments were performed at Ib boutons in muscles 6 and 7. (Figure 1A)
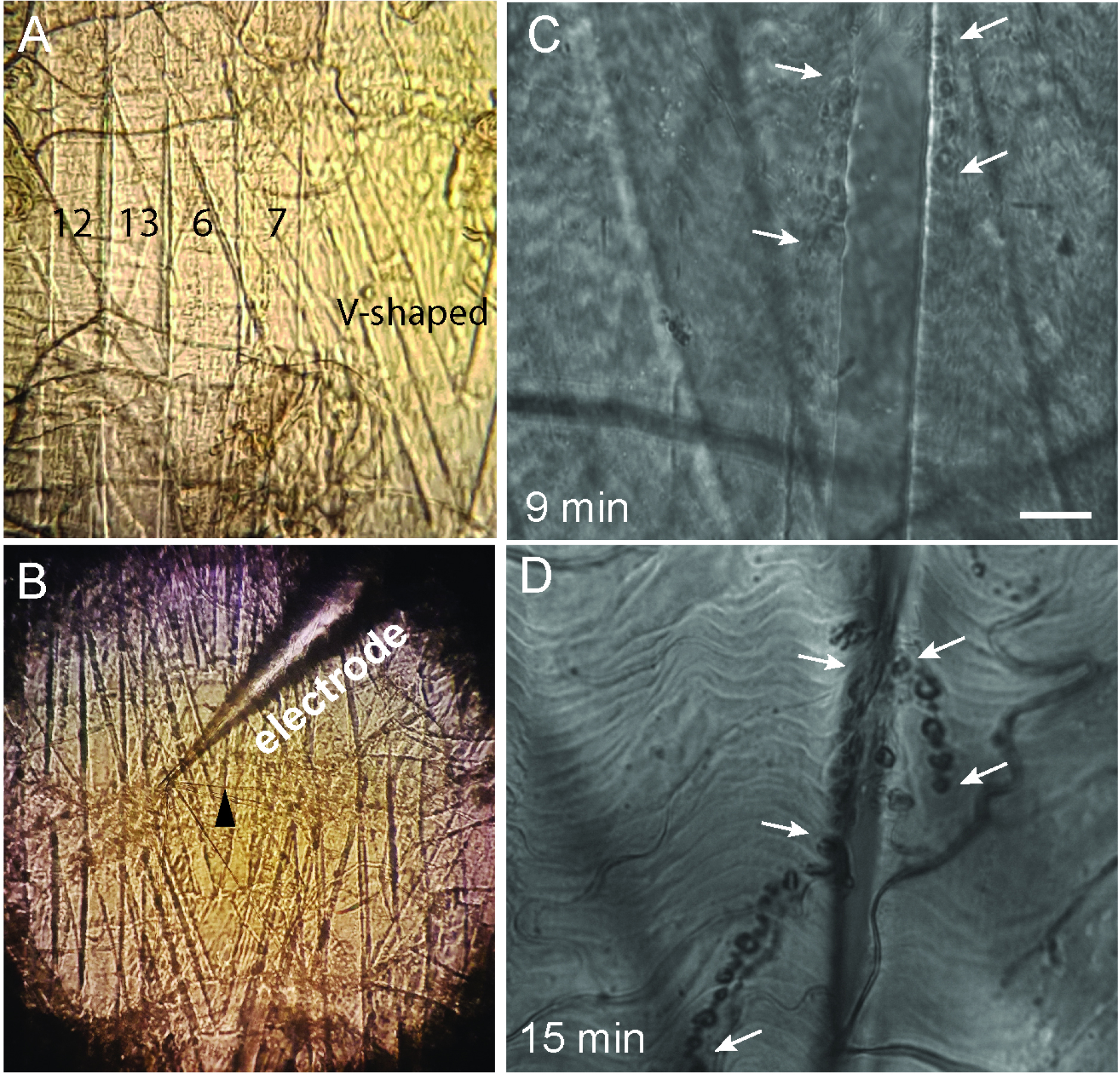
Figure 1. Third-instar larvae dissection. A. High magnification image of muscle fibers 7, 6, 13, 12 and V-shaped; B. Image of the ventral muscles in L3 where a nerve (black arrowhead) is sucked into the suction electrode connected to a stimulator; C and D. Adjusting the illumination times of the pre-fixed sample loaded with FM1-43 (WT larvae loaded for 30 sec). C. The DIC image shows that the illumination time of 9 min produces a successful photoconversion. Synaptic boutons can be identified as dark spots barely visible in the surface of the muscle (white arrows). D. The illumination time of 15 min is excessive, as evident from strong DAB precipitation, observed as large dark spots over the length of the NMJ (white arrows). Scale bar = 10 µm.
- Select third-instar larvae (L3) and place it in a Sylgard dish.
- Activity dependent labeling of Synaptic Vesicles with FM1-43 dye
Note: Perform the following steps in low light conditions to protect the fluorophore component of FM1-43 dye. Cover the FM1-43 dye with foil or work in the dark at room temperature.- Replace HL3 solution with HL3 containing 10 µM FM1-43.
- For loading in the absence of stimulation (spontaneous recycling pool) leave the preparation for the time of loading (10, 30, 120, and 600 sec were used in our studies).
- For active loading, using the head stage controllers pull the nerve into a suction electrode connected to a stimulator. Suck the end of the nerve. Use the stimulation protocol for low-frequency stimulation, 5 min, 5 Hz. Quickly remove the FM1-43 dye once the stimulation is complete (Figure 1B). The technique as adopted from Verstreken et al., 2008.
- Upon dye loading, rapidly wash the preparations three times for 30 sec in Ca2+-free HL3 saline containing 75 µM Advasep-7.
- Replace HL3 solution with HL3 containing 10 µM FM1-43.
- Fixation
- Prefix the preparation (Recipe 2) for 15 min at room temperature.
- Wash three times for 5 min in HEPES buffer saline (Recipe 3).
- Leave the preparation covered in 100 mM NH4Cl for 10 min to quench autofluorescence of the fixative.
- Wash out the NH4Cl solution with HEPES buffer 2 times for 5 min.
- Prefix the preparation (Recipe 2) for 15 min at room temperature.
- FM1-43 photoconversion
- Preincubate the preparation for 10 min in HEPES buffer containing 1.5 mg/ml DAB.
- Place the sample under an epifluorescent microscope. Identify the area of interest under 60x water immersion objective, at room temperature.
- Illuminate the sample for 8-10 min using a mercury lamp with a 480 ± 10 bandpass excision filter using the maximum light intensity. The dye will bleach with time. Observe the process of photoconversion by switching periodically to the bright field. As photoconversion takes place, dark brown DAB precipitate localizes at the illuminated area. The illumination time needs to be adjusted, as illustrated in Figures 1C and 1D.
- Preincubate the preparation for 10 min in HEPES buffer containing 1.5 mg/ml DAB.
- Post-fixation
- Wash the sample in HEPES three times 5 min.
- Immerse the preparation in the fixative solution (Recipe 4) and fix the sample in BioWave for 2 min at 100 W. Place the temperature insert close to the preparation to monitor temperature fluctuations. Do not allow the temperature to exceed 25-26 °C.
- Unpin the preparation carefully from the Sylgard dish. Transfer the preparation into a sample bottle with a snap cap.
- Wash the sample in 90 mM sodium cacodylate buffer, pH 7.4 twice for 5 min.
- Wash the sample in HEPES three times 5 min.
- EM sample processing
Note: The following steps (unless specified) are performed at room temperature.- Osmium tetroxide post-fixation and uranyl acetate staining
- Incubate the preparation in 1% OsO4 solution for 1 h at room temperature. The unused solution can be stored refrigerated for up to 1 month. Perform all steps under a fume hood, using gloves and long sleeve lab coat. Use 500 µl of this solution to immerse the sample. (Note 4)
- Wash with 90 mM cacodylate buffer for 5 min.
- Wash with deionized water for 5 min.
- Incubate the preparation in 2% uranyl acetate for 30-60 min. The unused solution can be stored refrigerated for up to 2 weeks.
- Wash with deionized water for 5 min twice.
- Incubate the preparation in 1% OsO4 solution for 1 h at room temperature. The unused solution can be stored refrigerated for up to 1 month. Perform all steps under a fume hood, using gloves and long sleeve lab coat. Use 500 µl of this solution to immerse the sample. (Note 4)
- Dehydration
- Prepare separate solutions containing 50, 70, 90, and 100% of acetone.
- Add 1 ml of 50% acetone to the sample for 10 min.
- Add 1 ml of 70% acetone to the sample for 10 min.
- Add 1 ml of 90% acetone to the sample for 10 min.
- Add 1 ml of 100% acetone to the sample twice for 10 min.
- Prepare separate solutions containing 50, 70, 90, and 100% of acetone.
- Embedding
- Mix 2 volumes of embedding resin (Note 3) with one volume of 100% acetone. Add it to the preparation under continuous rotation for 1 h or overnight.
- Keep the sample in embedding resin for at least 18 h (overnight).
- Under a stereomicroscope, place the preparation over ACLAR film, and then identify and trim the area of interest.
- Place the preparation over a new ACLAR film with the muscles facing down and the cuticle facing up. Position an embedding capsule filled with polymerized resin upside-down on the top of the specimen. Polymerize at 60 °C for 24-36 h.
- Remove the ACLAR film. This will leave sample attached to the embedding capsule.
- Mix 2 volumes of embedding resin (Note 3) with one volume of 100% acetone. Add it to the preparation under continuous rotation for 1 h or overnight.
- Thin sectioning and EM imaging
- Remove the block from the embedding capsule and mount the block into a specimen holder of an ultramicrotome.
- Under a stereomicroscope, mark the area of interest with a razor blade. The marked area should be a trapezoid that would fit onto the slot grid.
- Trim the block over the outlined area.
- Insert specimen holder with the block in the ultramicrotome and align the long edge of the trapezoid to the trimming knife edge. Trim 15-20 µm and shift the knife to trim the second long edge of the trapezoid. Trim another 15-20 µm. Repeat the same for the short sides of the trapezoid (Figure 2 and Videos 1-5).
- Align the block face exactly parallel to the edge of the diamond sectioning knife.
- Cut thin sections (50-60 nm).
- Collect individual sections (or collect sections in series) and place them on the 2 x 1 mm slot grids.
- Allow the sections to dry before taking images.
- Image the sections using conventional TEM.
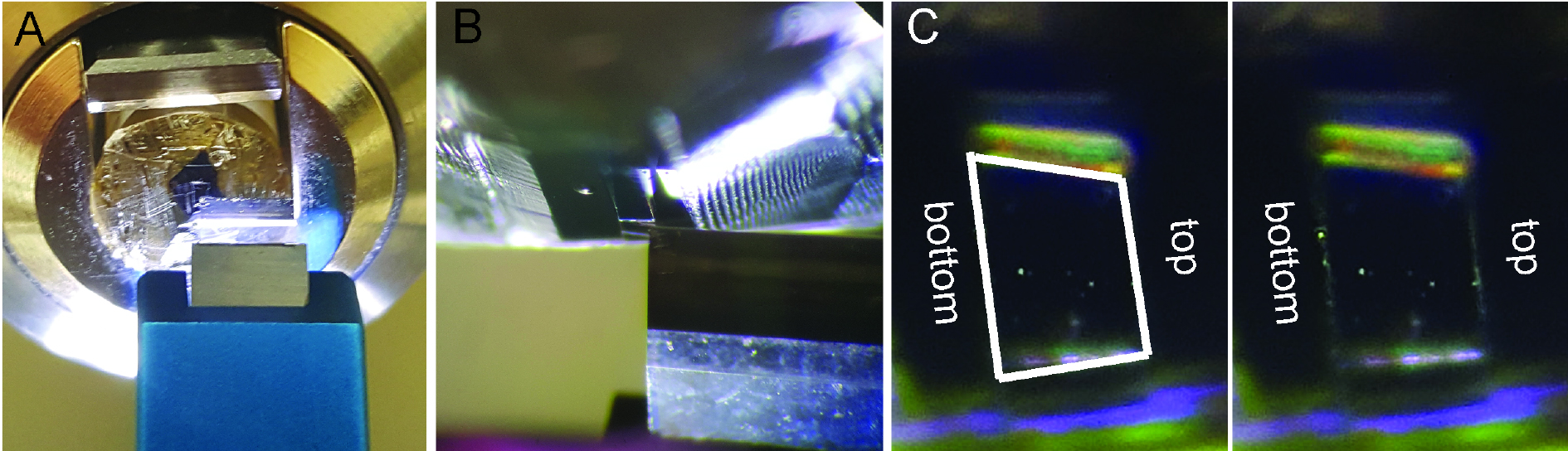
Figure 2. Sample block fine trimming. In the fixed and flat embedded Drosophila larvae the region of interest, muscles 6 and 7 are outlined with razor blade. Upon rough block trimming with razor blade and elimination of resin the face of the block is shaped with trimming knife. A. Fine trimming. The edge of the knife is aligned to the trapezoid top. B. Vertically positioned trapezoid with the left edge of the trimming knife aligned to the top of the trapezoid. C. Fine trimmed trapezoid with parallel top and bottom, and slightly tilted sides.
Note: The paralleled top and bottom are important for forming a ribbon during serial sectioning.Video 1. Rough trapezoid trimmingVideo 2. Fine trapezoid trimming, part one. Trimming trapezoid top.Video 3. Fine trapezoid trimming, part 2. Once sectioned the trapezoid top the trimming knife is retracted and positioned to trapezoid bottom without additional alignment.Video 4. Fine trapezoid trimming, part 3. Trimming trapezoid bottom.Video 5. Serial sectioning
- Osmium tetroxide post-fixation and uranyl acetate staining
Data analysis
- Employing the photoconversion procedure, we investigated the recycling vesicle pool in the Drosophila larvae lacking the presynaptic protein complexin (cpx-/-) (Huntwork and Figure 3A), which enables the elevated rate of spontaneous release. In contrast, the recycling pool observed in WT boutons during spontaneous loading was negligibly small (data not shown, see Sabeva et al., 2017). In cpx -/-preparations loaded during the nerve stimulation, the recycling pool was also increased compared to WT preparations (Figures 3A and 3B), suggesting that separate recycling pools are utilized for the spontaneous and evoked release modes (Sabeva et al., 2017).
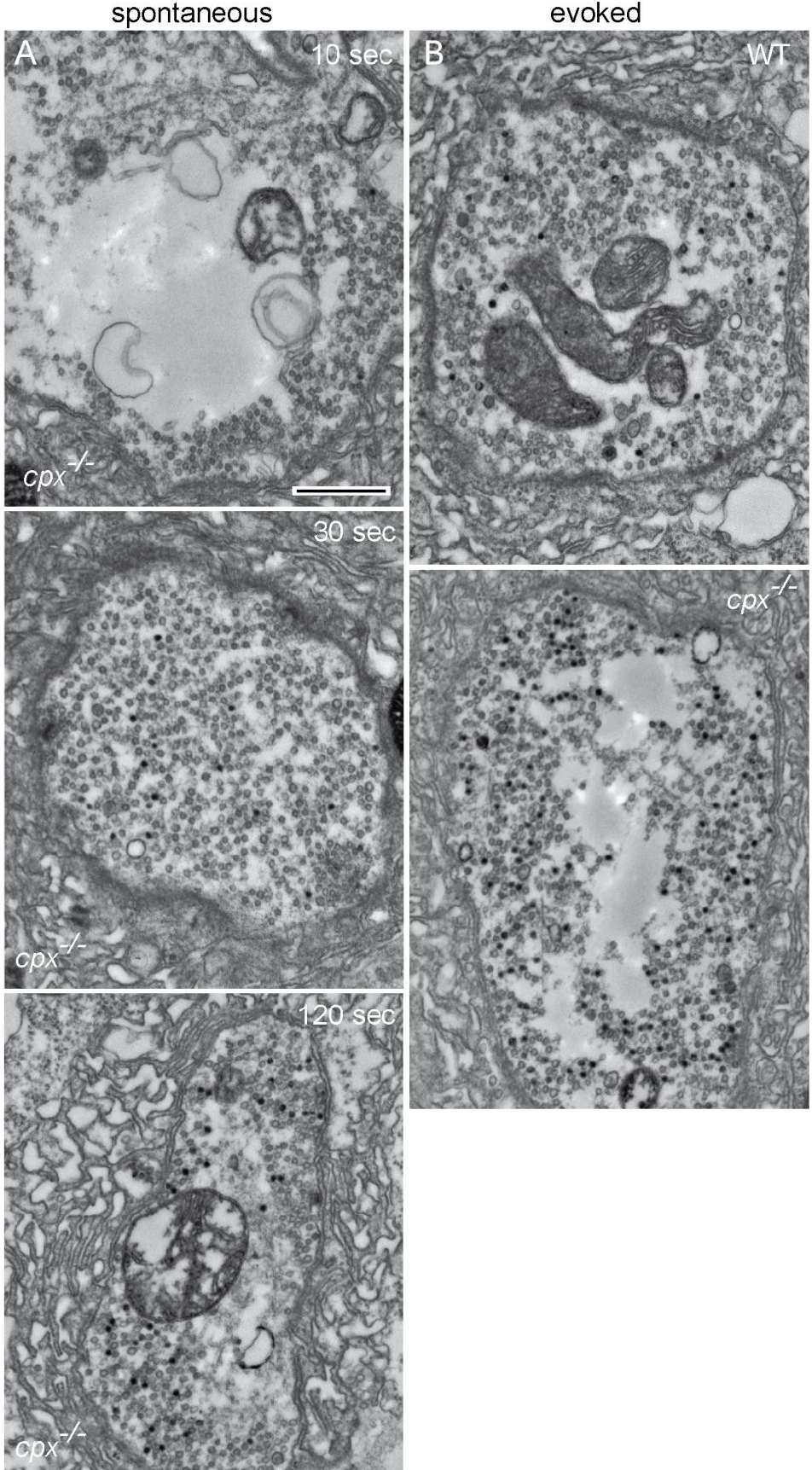
Figure 3. Representative micrographs showing the recycling vesicle pools. A. cpx-/- boutons loaded in the absence of stimulation for 10, 30 or 120 sec. B. WT and cpx-/- boutons loaded during the nerve stimulation at 5 Hz for 5 min. Photoconverted labelled vesicles (black), non-labelled vesicles (grey). Scale bar = 500 nm. - Detailed data processing analysis and replicates including applied statistical tests could be found in the original manuscript, see Sabeva et al., 2017.
Notes
- Advasep-7 is a β-cyclodextran derivative. It works as a dye scavenger and reduces background fluorescence.
- 16% paraformaldehyde solution and 25% glutaraldehyde solution can be purchased in 10 ml ampoules.
- Embedding resin: Prepare in a new disposable 250 ml beaker by mixing equal volumes of Embedding mix A (Recipe 5) and Embedding mix B (Recipe 6). Use a wooden tongue depressor to mix thoroughly. Degas (eliminate the air bubbles) for 5 min in a vacuum desiccator or leave 30 min at room temperature. Store tightly sealed at room temperature and use at the same day or aliquot and store at -20 °C.
- All steps during EM sample processing are performed in shaker or sample rotator.
Recipes
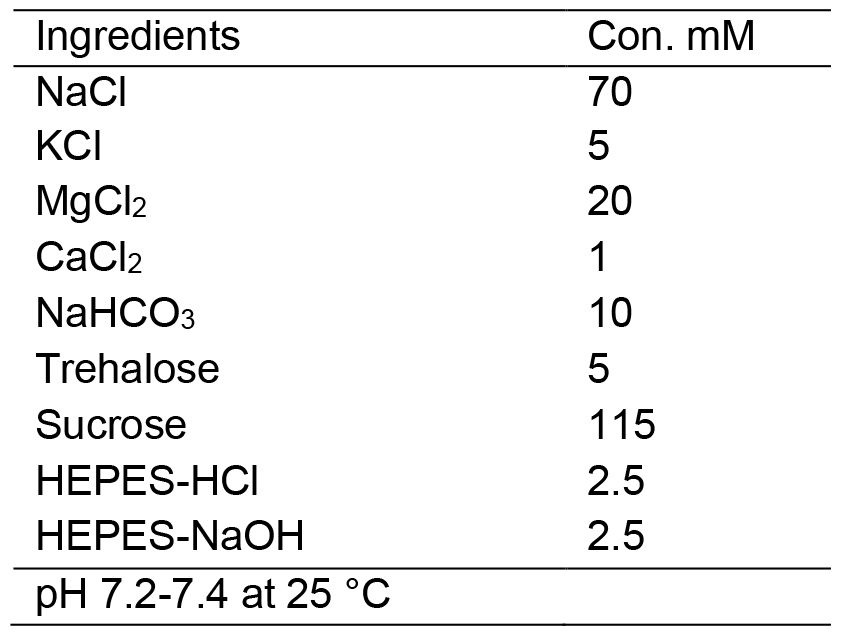
- HL3 Drosophila saline solution
- Pre-fixative solution
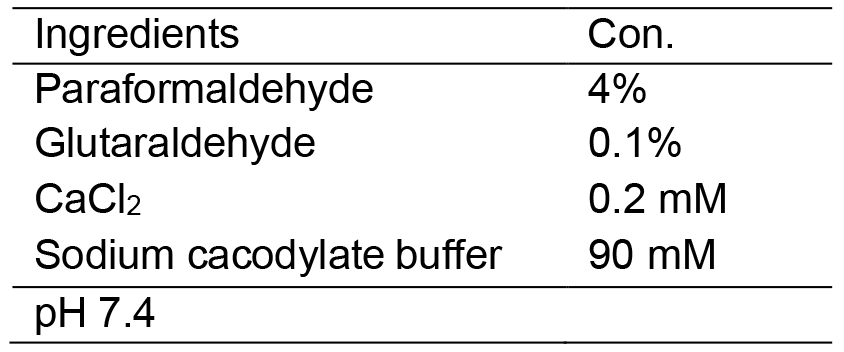
- HEPES-buffered saline

- Fixative solution
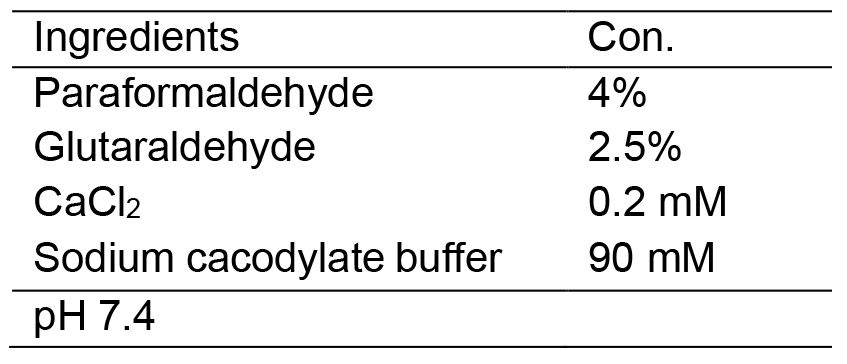
- Embedding mix A
Prepare by mixing:
20 g EMBed-812
11 g DDSA
9 g NMA
0.8 ml DMP-30
In 250 ml Tri-Corn disposable beaker
Use a wooden tongue depressor to mix thoroughly - Embedding mix B
Prepare by mixing:
10 g ERL
6 g DER
26 g NSA
0.4 ml DMAE
In 250 ml Tri-Corn disposable beaker
Use a wooden tongue depressor to mix thoroughly
Acknowledgments
This work was supported by National Institutes of Health Grant R01 MH099557 and NIH NINDS SNRP Grant 5U54NS083924.
References
- Akbergenova, Y. and Bykhovskaia, M. (2009). Enhancement of the endosomal endocytic pathway increases quantal size. Mol Cell Neurosci 40(2): 199-206.
- Denker, A., Krohnert, K. and Rizzoli, S. O. (2009). Revisiting synaptic vesicle pool localization in the Drosophila neuromuscular junction. J Physiol 587(Pt 12): 2919-2926.
- Harata, N., Ryan, T. A., Smith, S. J., Buchanan, J. and Tsien, R. W. (2001). Visualizing recycling synaptic vesicles in hippocampal neurons by FM 1-43 photoconversion. Proc Natl Acad Sci U S A 98(22): 12748-12753.
- Huntwork, S. and Littleton, J. T. (2007). A complexin fusion clamp regulates spontaneous neurotransmitter release and synaptic growth. Nat Neurosci 10(10): 1235-1237.
- Kavalali, E. T. (2006). Synaptic vesicle reuse and its implications. Neuroscientist 12(1): 57-66.
- Rizzoli, S. O. and Betz, W. J. (2004). The structural organization of the readily releasable pool of synaptic vesicles. Science 303(5666): 2037-2039.
- Rizzoli, S. O. and Betz, W. J. (2005). Synaptic vesicle pools. Nat Rev Neurosci 6(1): 57-69.
- Sabeva, N., Cho, W. R., Vasin, A., Gonzalez, A., Littleton, T. J. and Bykhovskaia, Maria. (2017). Complexin mutants reveal partial segregation between recycling pathways that drive evoked and spontaneous neurotransmission. J Neurosci 37: 383-396.
- Schikorski, T. and Stevens, C. F. (2001). Morphological correlates of functionally defined synaptic vesicle populations. Nat Neurosci 4(4): 391-395.
- Slepnev, V. I. and De Camilli, P. (2000). Accessory factors in clathrin-dependent synaptic vesicle endocytosis. Nat Rev Neurosci 1(3): 161-172.
- Sudhof, T. C. (2004). The synaptic vesicle cycle. Annu Rev Neurosci 27: 509-547.
- Verstreken, P., Ohyama, T. and Bellen, H. J. (2008). FM 1-43 labeling of synaptic vesicle pools at the Drosophila neuromuscular junction. Methods Mol Biol 440: 349-369.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Sabeva, N. and Bykhovskaia, M. (2017). FM1-43 Photoconversion and Electron Microscopy Analysis at the Drosophila Neuromuscular Junction. Bio-protocol 7(17): e2523. DOI: 10.21769/BioProtoc.2523.
- Sabeva, N., Cho, W. R., Vasin, A., Gonzalez, A., Littleton, T. J. and Bykhovskaia, Maria. (2017). Complexin mutants reveal partial segregation between recycling pathways that drive evoked and spontaneous neurotransmission. J Neurosci 37: 383-396.
Category
Neuroscience > Development > Neuron
Cell Biology > Cell imaging > Electron microscopy
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link











