- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Measurements of Free-swimming Speed of Motile Salmonella Cells in Liquid Media
Published: Vol 7, Iss 1, Jan 5, 2017 DOI: 10.21769/BioProtoc.2093 Views: 9615
Reviewed by: Arsalan DaudiKanika GeraAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Analysis of Gram-negative Bacteria Peptidoglycan by Ultra-performance Liquid Chromatography
Laura Alvarez [...] Felipe Cava
Oct 5, 2020 5450 Views

Assessing Swarming of Aerobic Bacteria from Human Fecal Matter
Arjun S. Byju [...] Sridhar Mani
May 5, 2021 3903 Views

High-speed Atomic Force Microscopy Observation of Internal Structure Movements in Living Mycoplasma
Kohei Kobayashi [...] Makoto Miyata
Mar 5, 2022 3004 Views
Abstract
Bacteria such as Escherichia coli and Salmonella enterica swim in liquid media using the bacterial flagella. The flagellum consists of the basal body (rotary motor), the hook (universal joint) and the filament (helical screw). Since mutants with a defect in flagellar assembly and function cannot swim smoothly, motility assay is an easy way to characterize flagellar mutants. Here, we describe how to measure free-swimming speeds of Salmonella motile cells in liquid media. Free-swimming behavior under a microscope shows a significant variation among bacterial cells.
Keywords: Bacterial flagellaBackground
The flagellar motor of E. coli and Salmonella is powered by downhill proton translocation along proton motive force (PMF) across the cytoplasmic membrane (Morimoto and Minamino, 2014; Minamino and Imada, 2015). The rotational speed of the proton-driven flagellar motor is proportional to total PMF (Gabel and Berg, 2003). Therefore, measurements of free-swimming speeds of motile cells allow us not only to analyze motor performance of various mutants but also to examine whether there is a significant difference in total PMF under experimental conditions (Minamino et al., 2016).
Materials and Reagents
- 1.5 ml Eppendorf tubes
- Double-sided tape (NICHIBAN, catalog number: NW-5 )
- Glass slide (Matsunami Glass, catalog number: S1126 )
- 18 x 18 mm coverslip (thickness: 0.12-0.17 mm) (Matsunami Glass, catalog number: C018181 )
- Pipette tips
- Filter paper
- Salmonella SJW1103 strain (wild type for motility and chemotaxis) (Yamaguchi et al., 1984)
- Salmonella MMHI0117 strain [∆fliH-fliI flhB(P28T)] (Minamino and Namba, 2008)
- Bacto tryptone (BD, catalog number: 211705 )
- Potassium dihydrogenphosphate (Wako Pure Chemical Industries, catalog number: 164-22635 )
- Dipotassium hydrogenphosphate (Wako Pure Chemical Industries, catalog number: 164-04295 )
- Bacto yeast extract (BD, catalog number: 212750 )
- Bacto agar (BD, catalog number: 214010 )
- Sodium chloride (Wako Pure Chemical Industries, catalog number: 192-13925 )
- T-broth (TB) (see Recipes)
- L-broth agar plate (see Recipes)
Equipment
- Selection of single channel pipettes (1,000 µl, 100 µl) (Gilson, model: P-1000 , P-100 )
- Shaking incubator (30 °C, at 200 rpm) (TAITEC, model: BR-40LF )
- Centrifuge (able to hold 1.5 ml tube, spin at 6,000 x g) (TOMY SEIKO, model: MX-305 )
- Spectrophotometer (able to measure OD600) (GE Healthcare, model: GeneQuant 1300 )
Note: This product has been discontinued by the manufacturer. - Phase contrast microscope (Olympus, model: CH40 )
- 40x objective lens
- CCD camera (Hamamatsu Photonics, model: C5405 )
- Objective micrometer (10 µm/pitch)
- Hard-disk video recorder (Panasonic, model: DMR-XP25V )
Software
- Move-tr/2D (Library Co., Tokyo)
- Microsoft Excel (Microsoft)
Procedure
Note: Carry out procedures at ca. 23 °C unless otherwise specified.
- Pick a single colony from L-broth agar plate and inoculate it into 5 ml of fresh TB.
- Incubate overnight at 30 °C with shaking at 150 rpm.
- Inoculate 50 µl of overnight culture of Salmonella SJW1103 and MMHI0117 strains into 5 ml of fresh TB and incubate at 30 °C for 5 h with shaking at 150 rpm. (The cell density reaches an OD600 of ca. 1.0-1.2.)
- Transfer 100 µl of the culture into a 1.5 ml Eppendorf tube.
- Collect the cells by centrifugation (6,000 x g, 2 min).
- Suspend the cell pellet in 1.0 ml of fresh TB.
- Centrifuge at 6,000 x g for 2 min.
- Discard supernatant.
- Repeat steps 4-6.
- Resuspend the cells in 1.0 ml of fresh TB.
- Make a tunnel slide by sandwiching double-sided tape between glass slide (bottom side) and 18 x 18 mm coverslip (top side) (see Figure 1 and Video 1).
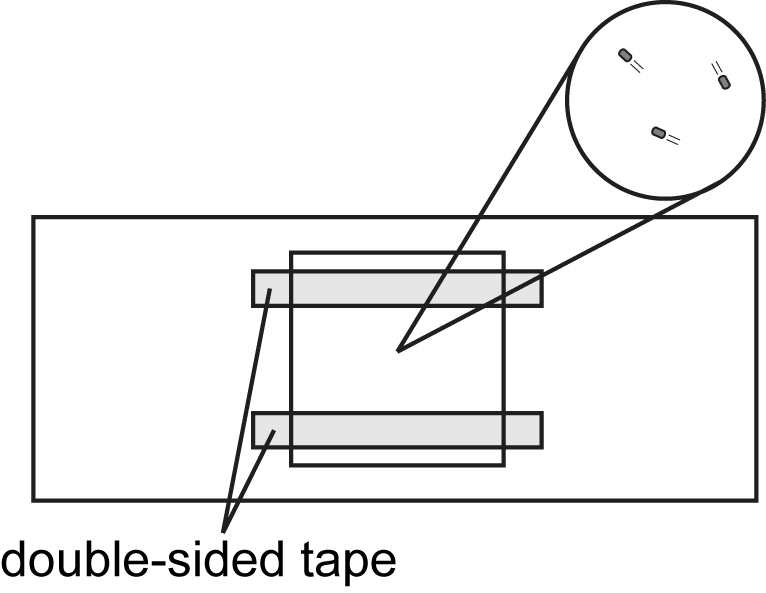
Figure 1. Tunnel slide for observation of bacteria cells under optical microscopy. Double-sided tapes are sandwiched between a glass slide (bottom side) and an 18 x 18 mm coverslip (top side). Cells are added to the space between the glass slide and the coverslip. Observation area is magnified in a circle.
Video 1. Preparation for a tunnel slide - Add the cell suspension to the tunnel slide and absorb excess medium with a piece of filter paper.
- Set the tunnel slide on the stage of a phase contrast microscope.
- Select a 40x objective lens.
- Observe motile cells under the phase contrast microscope.
Video 2. Free-swimming of Salmonella cells - Record a movie of motile cells for 10-30 sec with a hard-disk video recorder.
- Move a field of view under the microscope.
- Repeat steps 14 and 15 more than 5 times.
- Capture an image of an objective micrometer in the same setting.
- Tansfer the movies recorded on the hard-disk video recorder to a DVD.
Data analysis
- Calibrate a scale of a movie using the image of the objective micrometer.
- Track motile cells in the movie with the length of 1 sec (30 sequential images recorded at 30 frames per second) using a motion analysis software Move-tr/2D (Library Co., Tokyo).
- Measure the velocity (µm/sec) of each cell by the Move-tr/2D.
- Calculate the average velocity and standard deviation from the data of more than 30 cells using Microsoft Excel (Microsoft).
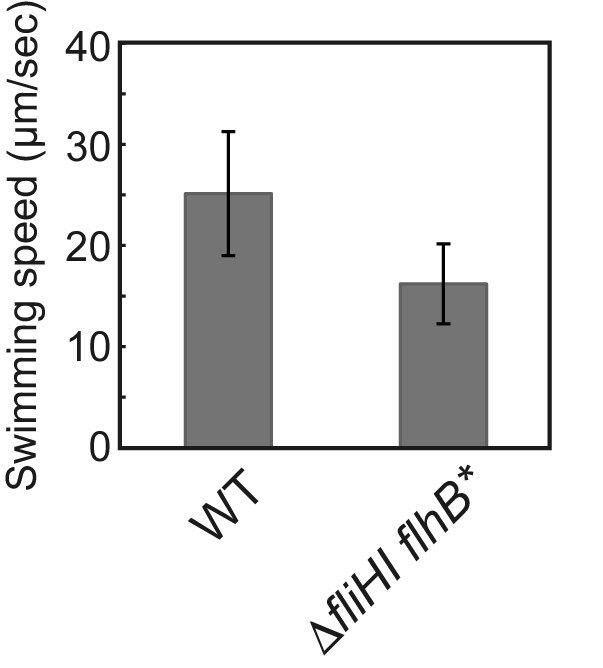
Figure 2. Free-swimming speed of Salmonella cells. SJW1103 (WT) and MMHI0117 (∆fliHI flhB*) were grown at 30 °C for 4 h in T-broth and observed by a phase contrast microscope. Vertical bars indicate standard deviations. (Modified from Minamino et al., 2016)
Notes
- When free swimming of motile cells is observed by phase contrast microscopy, a proper cell concentration is required. If the cell concentration is low, only a small number of cells are observed in a single movie. If the cell concentration is quite high, traces of each motile cell are overlapped, thereby making precise cell tracking difficult. The free-swimming motility is not affected at all by cell-cell interactions.
- Cells nonspecifically attached on a glass surface are often observed and so cannot show any free-swimming motility. So these cells must not be judged as non-motile cells.
Recipes
- T-broth (TB)
1% Bacto tryptone, 10 mM potassium phosphate, pH 7.5 - L-broth agar plate
1% Bacto tryptone, 0.5% Bacto yeast extract, 1% NaCl, 1.5% Bacto agar
Acknowledgments
This protocol was modified from a previous work (Minamino et al., 2003). This research has been supported in part by JSPS KAKENHI Grant Numbers JP15K14498 and JP15H05593 to YVM, JP21227006 and JP25000013 to KN and JP26293097 to TM and MEXT KAKENHI Grant Numbers JP26115720 and JP15H01335 to YVM and JP23115008, JP24117004, JP25121718 and JP15H01640 to TM.
References
- Gabel, C. V. and Berg, H. C. (2003). The speed of the flagellar rotary motor of Escherichia coli varies linearly with protonmotive force. Proc Natl Acad Sci USA 100(15): 8748-8751.
- Minamino, T. and Imada, K. (2015). The bacterial flagellar motor and its structural diversity. Trends Microbiol 23(5): 267-274.
- Minamino, T., Imae, Y., Oosawa, F., Kobayashi, Y. and Oosawa, K. (2003) Effect of intracellular pH on rotational speed of bacterial flagellar motors. J Bacteriol 185(4):1190-1194.
- Minamino, T., Morimoto, Y. V., Hara, N., Aldridge, P.D. and Namba, K. (2016). The bacterial flagellar type III export gate complex is a dual fuel engine that can use both H+ and Na+ for flagellar protein export. PLoS Pathog 12(3): e1005495.
- Minamino, T. and Namba, K. (2008). Distinct role of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature 451(7177): 485-488.
- Morimoto, Y. V. and Minamino, T. (2014). Structure and function of the bi-directional bacterial flagellar motor. Biomolecules 4(1): 217-234.
- Yamaguchi, S., Fujita, H., Sugata, K., Taira, T. and Iino, T. (1984). Genetic analysis of H2, the structural gene for phase-2 flagellin in Salmonella. J Gen Microbiol 130(2): 255-265.
Article Information
Copyright
© 2017 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Readers should cite both the Bio-protocol article and the original research article where this protocol was used:
- Morimoto, Y. V., Namba, K. and Minamino, T. (2017). Measurements of Free-swimming Speed of Motile Salmonella Cells in Liquid Media. Bio-protocol 7(1): e2093. DOI: 10.21769/BioProtoc.2093.
- Minamino, T., Morimoto, Y. V., Hara, N., Aldridge, P.D. and Namba, K. (2016). The bacterial flagellar type III export gate complex is a dual fuel engine that can use both H+ and Na+ for flagellar protein export. PLoS Pathog 12(3): e1005495.
Category
Microbiology > Microbial cell biology > Cell-based analysis
Cell Biology > Cell movement > Cell motility
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link










