- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Antibiotic Disc Assay for Synechocystis sp. PCC6803
Published: Vol 6, Iss 24, Dec 20, 2016 DOI: 10.21769/BioProtoc.2071 Views: 11106
Reviewed by: Maria SinetovaElizabeth LibbyLaura Molina-García

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

SIMBA Method—Simultaneous Detection of Antimicrobial and Anti-biofilm Activity of New Compounds Using Salmonella Infantis
Meta Sterniša [...] Anja Klančnik
Aug 5, 2023 2016 Views

Functional Assay for Measuring Bacterial Degradation of Gemcitabine Chemotherapy
Serkan Sayin and Amir Mitchell
Sep 5, 2023 1743 Views

Identification of Mycobacterium tuberculosis and its Drug Resistance by Targeted Nanopore Sequencing Technology
Chen Tang [...] Guangxin Xiang
Feb 5, 2025 1992 Views
Abstract
This protocol describes how to investigate the integrity of the outer cell wall in the cyanobacterium Synechocystis sp. PCC6803 using antibiotics. It is adapted to the agar diffusion test (Bauer et al., 1966), in which filter paper discs impregnated with specified concentrations of antibiotics were placed on agar plates inoculated with bacteria. The antibiotics we tested, interfering with the biosynthesis/function of bacterial cell walls, will diffuse into the agar and produce a zone of cyanobacterial growth inhibition around the disc(s). The size of the inhibition zone reflects the sensitivity of the strain to the action of antibiotics, e.g., a mutation in a protein functioning within the cell wall or its construction would render the mutant strain more sensitive to the respective antibiotic. The method has proven to be useful for phenotyping a mutant of Synechocystis sp. PCC6803 lacking all three genes encoding Deg proteases. Deletion of these ATP-independent serine proteases was shown to have impact on the outer cell layers of Synechocystis cells (Cheregi et al., 2015).
Keywords: CyanobacteriaBackground
The cyanobacterium Synechocystis sp. PCC6803 (hereafter, Synechocystis 6803) is a model organism for studying the process of photosynthesis. While its genome was sequenced already in 1996, still more than 50% of its genes encode proteins with hypothetical or unknown function. The three genes slr1204 (htrA), sll1679 (hhoA) and sll1427 (hhoB) encode serine proteases of the Deg (degradation of periplasmic proteins) family; despite detailed analyses (see Cheregi et al., 2016 and references therein) their exact subcellular localization and substrates still are enigmatic. Previous proteomic and metabolomic characterizations of single and triple deg deletion mutants performed in our lab have shown altered expression of proteins with functions in or on the outer cell layers of Synechocystis 6803 (Miranda et al., 2013; Tam et al., 2015; Cheregi et al., 2015).
The antibiotics carbenicillin, colistin and polymyxin inhibit or disrupt the bacterial cell wall and therefore can be used to test the integrity of this cellular component in mutants: polymyxin acts on the outermost lipopolysaccharide layer surrounding the cyanobacterial S-layer, carbenicillin interferes with the peptidoglycan layer and colistin acts on the plasma membrane of gram-negative bacteria (Table 1). An agar diffusion test has been developed (Bauer et al., 1966) in which filter paper discs impregnated with specified concentrations of antibiotics are placed on agar plates inoculated with bacteria. The antibiotics will diffuse from the disc into the agar and inhibit cyanobacterial growth around it. The size of this inhibition zone then reflects the sensitivity of the strain to the antibiotic. The antibiotic disc assay method was used to characterize a triple deg protease mutant, and could be used for the characterization of any cyanobacterial mutant. However, the reader should be aware of the limitations of this assay. Despite the Sll1951 protein being the main component of the outermost cell layer of cyanobacteria, called S-layer, the antibiotic disc assay only had limited effect on a sll1951 deletion mutant (Trautner and Vermaas, 2013). Though the S-layer is compromised in the sll1951 deletion mutant, the underlying layers are still intact, preventing Carbenicillin and Polymyxin B, due to their relatively high molecular masses, to penetrate into the cell.
Table 1 describes the molecular weight, the mode of action and the ordering information for the above mentioned antibiotic discs.
Table 1. Antibiotic discs used in this protocol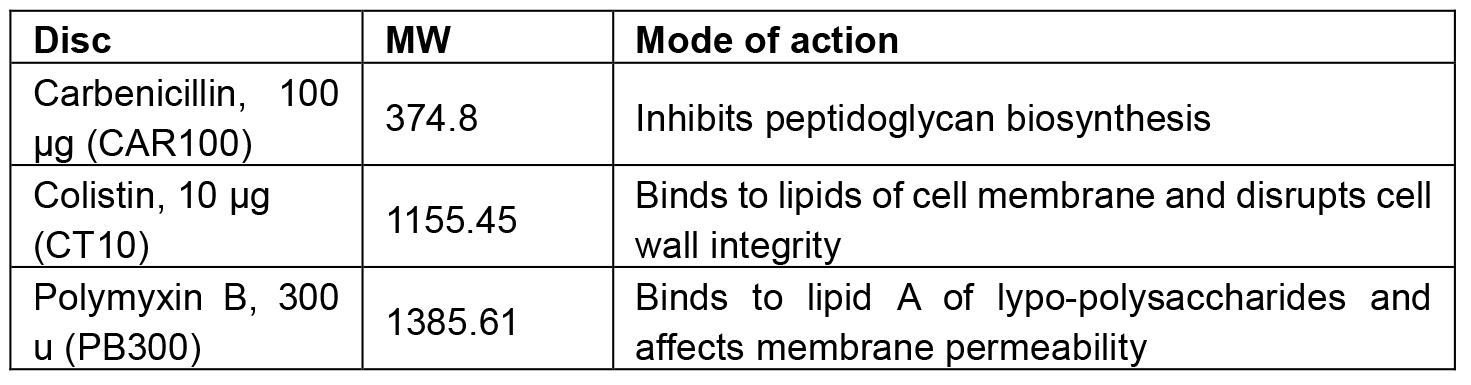
Materials and Reagents
- RemelTM plastic Petri dishes (85 mm diameter) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: R80085 )
- Sterile polystyrene spreading rods (SARSTEDT, catalog number: 86.1569.005 )
- Carbenicillin (CAR100) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: CT0006B )
- Colistin (CT10) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: CT0017B )
- Polymyxin (PB300) (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: CT0044B )
- Dispenser for discs (included in the kit of each of the above mentioned test discs)
- Cyanobacterial cultures of WT control and mutants to be tested
- Boric acid, H3BO3 (Sigma-Aldrich, catalog number: B6768 )
- Manganese(II) chloride tetrahydrate, MnCl2·4H2O (Sigma-Aldrich, catalog number: 221279 )
- Zinc sulfate heptahydrate, ZnSO4·7H2O (Sigma-Aldrich, catalog number: Z1001 )
- Sodium molybdate dehydrate, Na2MoO4·2H2O (Sigma-Aldrich, catalog number: 331058 )
- Cupric sulfate pentahydrate, CuSO4·5H2O (Thermo Fisher Scientific, Fischer Scientific, catalog number: C493-500 )
- Cobalt(II) nitrate hexahydrate, Co(NO3)2·6H2O (Sigma-Aldrich, catalog number: 239267 )
- Sodium nitrate, NaNO3 (Scharlab, catalog number: SO05010500 )
- Magnesium sulfate heptahydrate, MgSO4·7H2O (Sigma-Aldrich, catalog number: 63138 )
- CaCl2·2H2O (Scharlab, catalog number: CA01981000 )
- Citric acid (Sigma-Aldrich, catalog number: 251275 )
- Na2-EDTA (Sigma-Aldrich, catalog number: 27285 )
- Ferric ammonium citrate (Sigma-Aldrich, catalog number: F5879 )
- Sodium carbonate, Na2CO3(Sigma-Aldrich, catalog number: 71345 )
- di-potassium hydrogen phosphate, K2HPO4 (EMD Millipore, catalog number: 105104 )
- Na-thiosulfate (solid) (Sigma-Aldrich, catalog number: 217263 )
- Difco Bacto-agar (BD, catalog number: 214530 )
- TES (Sigma-Aldrich, catalog number: T6541 )
- 100x BG11 without Fe, phosphate, carbonate (see Recipes)
- 1,000x ferric ammonium citrate (see Recipes)
- 1,000x Na2CO3 (see Recipes)
- 1,000x K2HPO4 (see Recipes)
- BG11 solid agar plates (see Recipes)
- 1 M TES/NaOH buffer, pH 8.2 (see Recipes)
Equipment
- Cell culture flasks (50-250 ml) with vented caps (TC flask T25) (SARSTEDT, catalog number: 83.3910.002 )
- UV/VIS spectrophotometer (GlobalMarket, PG Instruments, model: T90+ ) for measuring the absorption of cell culture (OD730).
- MultisizerTM Coulter Counter for counting cells (Beckman Coulter, model: Z Series Coulter Counter )
- Laminar hood (Thermo Fisher Scientific, Thermo ScientificTM, model: HeraguardTM Eco Clean Bench )
- Shaking incubator (80-120 rotations/min) with light (IVIS ~60-100 µE m-2 s-1) and temperature adjusted to 30 °C (Eppendorf, model: New BrunswickTM Innova® 43 )
Procedure
Note: it is recommended that handling of cyanobacterial cultures is done under aseptic conditions using a sterile laminar hood.
- 100 ml of BG11 media are inoculated with cyanobacterial cells of WT and mutant at an initial OD730 ~0.05 and let to grow for 2-3 days until the OD of the cultures measured with the spectrophotometer at 730 nm is between 0.4-0.8.
- Count cells using the Beckman Coulter cell counter. Alternatively, if a cell counter is not available, cells can be counted using a hemocytometer.
- Working under sterile conditions, dilute the cyanobacterial cultures with BG11 to 100,000 cells/ml.
- Spread 1 ml of diluted cultures (containing 100,000 cells) of the WT and mutant strains on Petri dishes containing 40 ml of solid BG11 media respectively. Leave the plates open in the sterile hood until the liquid has evaporated.
- One-three antibiotic disks are placed on each plate of the WT and mutant strains. Use at least three biological replicates of each strain to be tested.
- Petri dishes are placed in an incubator with light and 30 °C and left to incubate (1-2 weeks) until green colonies are visible.
- Measure the inhibition area around each antibiotic disc to the last mm (Figure 1). If the mutant shows a higher zone of inhibition compared with the WT, this phenotype could be the consequence of the mutation interfering with the biogenesis/function of cell wall.
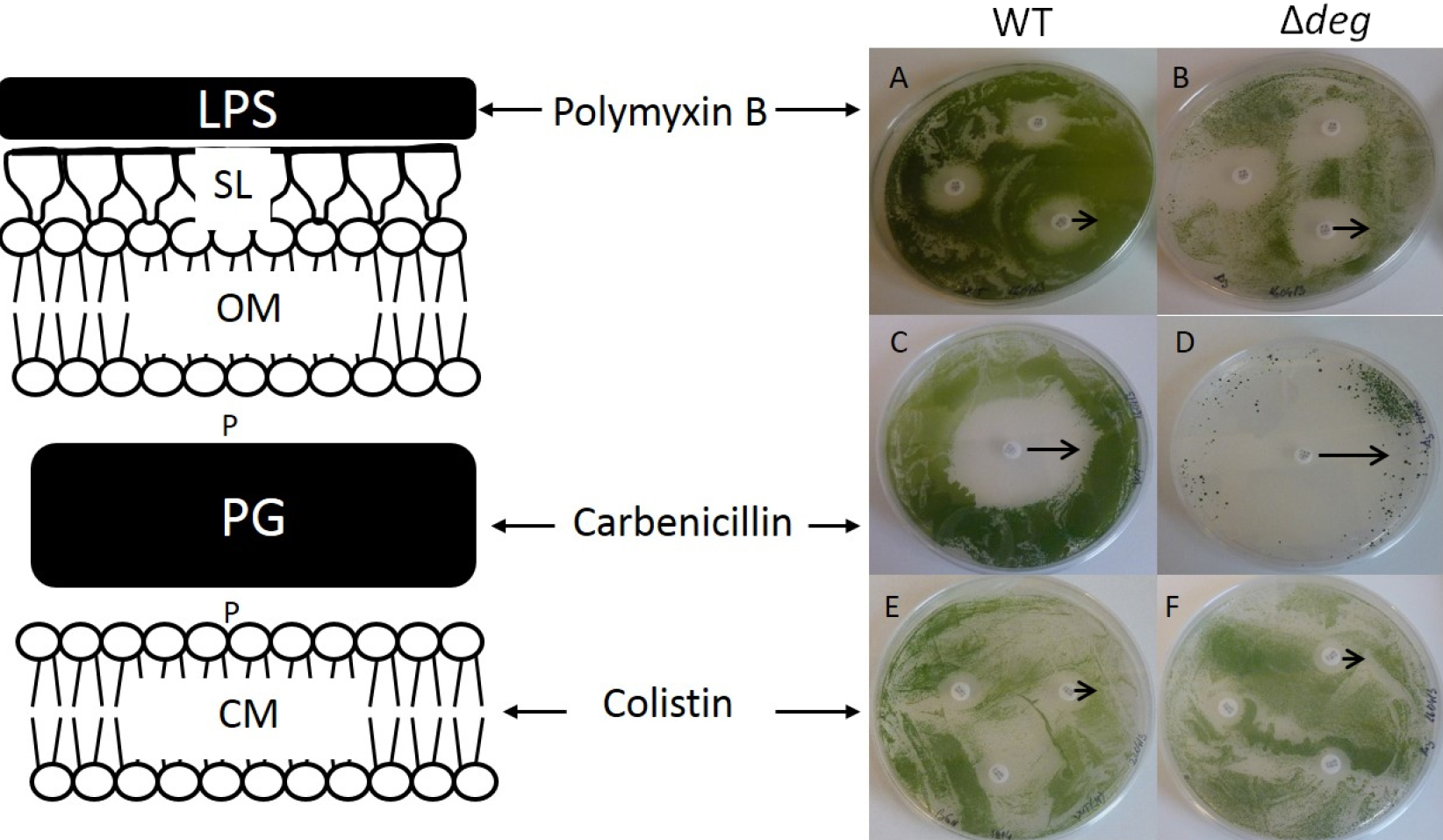
Figure 1. Antibiotic sensitivity test assay. Susceptibility of the WT (A, C, E) and Δdeg (B, D, F) to the different antibiotics is shown by their zone of inhibition (ZOI). ZOI induced by Polymixin B is 0.55 (± 0.1) cm and 0.8 (± 0.08) cm in WT (A) and Δdeg mutant (B) respectively. ZOI of Carbenicillin was 1.6 (± 0.19) cm for the WT (C) and 2.4 (± 0.25) cm for Δdeg (D). Colistin affected similarly WT (E) and Δdeg (F), ZOI of 0.4 cm. The black arrows indicate the measured zone of inhibitions. Adapted from Cheregi et al. (2015).
Data analysis
The average diameters of the areas of inhibition (in cm) and standard deviations were calculated from four biological replicates. Each biological replicate was represented by 3 technical replicates; the technical replicates presented areas of inhibition that were identical. These data are presented in Table 3 and Figure 3 of Cheregi et al. (2015).
Notes
This protocol was adapted from the previously published study of Trautner and Vermaas (2013) and it was performed as in Cheregi et al. (2015). The efficiency of the antibiotic is decreasing after expiration date.
Recipes
- Trace minerals (1 L)
2.86 g H3BO3
1.81 g MnCl2·4H2O
0.22 g ZnSO4·7H2O
0.39 g Na2MoO4·2H2O
0.079 g CuSO4·5H2O
0.049 g Co(NO3)2·6H2O - 100x BG11 without Fe, phosphate, carbonate (1 L)
149.6 g NaNO3
7.5 g MgSO4·7H2O
3.6 g CaCl2·2H2O
0.60 g citric acid (or 0.89 g Na-citrate, dihydrate)
1.12 ml 0.25 M Na2-EDTA, pH 8.0
100 ml trace minerals - Other components
Ferric ammonium citrate, 6 mg/ml (1,000x): 600 mg per 100 ml dH2O
Na2CO3 (1,000 x): 2 g Na2CO3 per 100 ml dH2O
K2HPO4 (1,000 x): 3.05 g K2HPO4 per 100 ml dH2O - BG11 liquid media (1 L)
10 ml 100x BG11 without Fe, phosphate, carbonate
1 ml 1,000x ferric ammonium citrate
1 ml 1,000x Na2CO3
1 ml 1,000x K2HPO4 - BG11 solid agar plates (1 L)
For agar plates, add to the above:
10 ml 1 M TES/NaOH buffer pH 8.2 (long term storage in the fridge)
3 g Na-thiosulfate (solid)
15 g Difco Bacto-agar
Autoclave at 121 °C for 30 min - 1 M TES/NaOH buffer, pH 8.2
Dissolve 229.2 gr of TES in distilled H2O
The resulting solution should be clear
Bring the pH to 8.2 using a concentrated solution of NaOH
Acknowledgments
This work was supported by grants from the Swedish Energy Agency (to C. Funk).
References
- Bauer, A. W., Kirby, W. M. M., Sherris, J. C. and Turck, M. (1966). Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol 45(4): 493-496.
- Cheregi, O., Miranda, H., Gröbner, G. and Funk, C. (2015). Inactivation of the Deg protease family in the cyanobacterium Synechocystis sp. PCC 6803 has impact on the outer cell layers. J Photoch Photobio B 152: 383-394.
- Cheregi, O., Wagner, R. and Funk, C. (2016). Insights into the cyanobacterial Deg/HtrA proteases. Front Plant Sci 7:624.
- Miranda, H., Cheregi, O., Netotea, S., Hvidsten, T. R., Moritz, T. and Funk, C. (2013). Co-expression analysis, proteomic and metabolomic study on the impact of a Deg/HtrA protease triple mutant in Synechocystis sp. PCC 6803 exposed to temperature and high light stress. J Proteomics 78: 294-311.
- Tam, L. X., Aigner, H., Timmerman, E., Gevaert, K. and Funk, C. (2015). Proteomic approaches to identify substrates of the three Deg/HtrA proteases of the cyanobacterium Synechocystis sp. PCC 6803. Biochem J 468(3): 373-384.
- Trautner, C. and Vermaas, W. F. (2013). The sll1951 gene encodes the surface layer protein of Synechocystis sp. strain PCC 6803. J Bacteriol 195(23): 5370-5380.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Cheregi, O. and Funk, C. (2016). Antibiotic Disc Assay for Synechocystis sp. PCC6803. Bio-protocol 6(24): e2071. DOI: 10.21769/BioProtoc.2071.
Category
Microbiology > Antimicrobial assay > Antibacterial assay
Microbiology > Microbial metabolism > Other compound
Cell Biology > Cell metabolism > Other compound
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link









