- Submit a Protocol
- Receive Our Alerts
- Log in
- /
- Sign up
- My Bio Page
- Edit My Profile
- Change Password
- Log Out
- EN
- EN - English
- CN - 中文
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
- EN - English
- CN - 中文
- Home
- Protocols
- Articles and Issues
- For Authors
- About
- Become a Reviewer
Fat Turnover Assay in Drosophila
Published: Vol 6, Iss 21, Nov 5, 2016 DOI: 10.21769/BioProtoc.1996 Views: 12483
Reviewed by: Masahiro MoritaLeonardo G. GuilgurAnonymous reviewer(s)

Protocol Collections
Comprehensive collections of detailed, peer-reviewed protocols focusing on specific topics
Related protocols

Measurement of 2-methylthio Modifications in Mitochondrial Transfer RNAs by Reverse-transcription Quantitative PCR
Fan-Yan Wei and Kazuhito Tomizawa
Jan 5, 2016 9526 Views

Quantification of 2-Hydroxyglutarate Enantiomers by Liquid Chromatography-mass Spectrometry
William M. Oldham and Joseph Loscalzo
Aug 20, 2016 11866 Views

A New Approach to Detect and Semi-quantify All Molecular Species and Classes of Anionic Phospholipids Simultaneously in Plant Samples
Manon Genva [...] Laetitia Fouillen
Apr 20, 2025 1723 Views
Abstract
Like all animals, Drosophila shows robust fat (triglyceride) turnover, i.e., they synthesize, store and utilize triglyceride for their daily metabolic needs. The protocol describes a simple assay to measure this turnover of triglycerides in Drosophila.
Background
Almost all animals store energy reserves in the form of glycogen and triglycerides. Many physiological, pathological and environmental conditions cause changes in the total level of these energy reserves, especially triglycerides. However, it’s not always clear whether the resulting changes in triglycerides are due to reduced breakdown, increased synthesis or vice versa. With this protocol, it is possible to determine both the rate of synthesis and degradation of the newly synthesized triglycerides in flies.
Materials and Reagents
- 1.5 ml Eppendorf tubes
- Hamilton glass syringes (Hamilton, catalog number: Gastight 1700 )
- Metallic needle
- Razor blades (VWR, catalog number: 55411 )
- TLC silica gel 60 plates (Figure S1) (EMD Millipore, catalog number: 105626 )
- Drosophila vial (Genesee Scientific, Flystuff, catalog number: 32-109 )
- Whatman® chromatography paper (Sigma-Aldrich, catalog number: WHA3030861 )
- Drosophila melanogaster
- D-[14C(U)]-glucose (PerkinElmer, catalog number: NEC042V250UC )
- Yeast extract
- Sugar
- Liquid nitrogen and nitrogen gas
- Chloroform (Sigma-Aldrich, catalog number: 366927 )
- Methanol (EMD Millipore, catalog number: MX0475-1 )
- Cupric sulfate acid (EMD Millipore, catalog number: 102790 )
- O-phosphoric acid (85%) (Thermo Fisher Scientific, Fisher Scientific, catalog number: A242-212 )
- Lipid standards:
- Triolein (Sigma-Aldrich, catalog number: T7140 )
- Phosphatidylcholine (Sigma-Aldrich, catalog number: P3556 )
- Phosphatidylinositol (Sigma-Aldrich, catalog number: P6636 )
- Cholesterol (Sigma-Aldrich, catalog number: C8667 )
- Lauric acid (Sigma-Aldrich, catalog number: L556 )
- Myristic acid (Sigma-Aldrich, catalog number: M3128 )
- Palmitic acid (Sigma-Aldrich, catalog number: P0500 )
- Hexane (Sigma-Aldrich, catalog number: 32293 )
- Diethyl ether (Sigma-Aldrich, catalog number: 309966 )
- Acetic acid (AMRESCO, catalog number: 0714 )
- Sodium chloride (NaCl) (Sigma-Aldrich, catalog number: S9888 )
- Scintillation fluid (PerkinElmer, catalog number: Ultima GoldTM/6013329 )
- Drosophila food recipes (see Recipes)
- Solvent mixture (see Recipes)
- Cupric sulfate/phosphoric acid solution (see Recipes)
- 0.9% NaCl solution (see Recipes)
Equipment
- TLC chamber (Clarkson Laboratory and Supply, model: Latch-Lid ChromatoTank 80-30 )
- Kontes microcentrifuge motor and pestles (Sigma-Aldrich, catalog number: Z359971-1EA )
- Centrifuge (Eppendorf, model: Centrifuge 5810R )
- Benchtop vacuum oven (VWR, model: 97027 )
- Scintillation counter (Beckman Coulter, model: LS6500 )
- Scintillation vials (PerkinElmer, catalog number: 6000292 )
- Nalgene PPCO wash bottles (Thermo Fisher Scientific, Thermo ScientificTM, catalog number: 2405-0500 )
Procedure
- The assay describes fat turnover in adult mated Drosophila females maintained on two different diets. The diets are HY (High yeast containing 5% yeast extract and 5% sugar) and LY (low yeast, containing 0.5% yeast extract and 5% sugar, see Katewa et al., 2016). On the tenth day about 300 flies (12 batches of 25 flies each) were transferred to fresh food vials with 2 µCi of 14C labeled glucose. Vials were prepared by adding 30 µl of 5% sugar/glucose solution on top and allowed to settle for 4 h. For preparing the sugar/glucose solution 100 µl of D-[14C(U)]-glucose was added to 900 µl of 5% sugar solution.
Note: When you add 30 µl of sugar/glucose solution on top of the food, it’s best to use the sugar concentration that is closer to the concentration of sugar present in the regular lab fly food. For example, if your lab uses 10% sugar in the fly food, use 10% sugar solution to make the sugar/glucose solution.
- The flies were maintained on the labeled food for 24 h, after which half of the flies were snap-frozen in liquid nitrogen. This is the 0 h sample. The other half was transferred to a fresh non-radioactive food vial and was kept on this food for the next 60 h, and was then immediately frozen. This is the 60 h sample.
- For lipid extraction, weigh about 20 mg of flies/replicate. The weight is important for normalization and extraction of lipids. The number of flies will differ depending on the diets used in the experiment. For example, to get about 20 mg of total weight from the HY group, I need about 15-18 female flies, whereas for the LY group, I need about 22-25 flies. Homogenize the flies in about 100 µl of 0.9% NaCl in a 1.5 ml Eppendorf tube with a Kontes microcentrifuge motor and pestle. Transfer the homogenate to a 2 ml glass vial. Use additional 100 µl of 0.9% NaCl solution to rinse the Eppendorf tube and transfer to the glass vial. Add 800 µl of chloroform:methanol (2:1, v/v) to the glass vial with the homogenate, vortex for 15 sec and let it stand at room temperature for 20 min. Vortex again for 15 sec and centrifuge at 1,640 x g for 10 min to separate the two phases. By using a glass Hamilton syringe, carefully remove the lower phase containing the lipid fractions (Folch et al., 1957).
- The lipid fraction is transferred to a glass vial and dried under a continuous stream of nitrogen gas (always in a chemical fume hood that is designated for radioactive work). A glass pipette or a metallic needle could be attached at the end of gas supply and suspended in the glass vials (about one inch away from the solvent surface). Once completely dried, add 100 μl of chloroform to the tube.
Note: There are two ways to separate the lipids into different fractions. One way is by using the solid phase extraction (SPE) tubes (for details see Katewa et al., 2012); another way is to use thin layer chromatography (TLC) plates (Chatterjee et al., 2014; Katewa et al., 2016). Here I am describing the TLC protocol as it is cost effective, simpler and also provides a visual separation of lipids.
- A glass Hamilton syringe is used to spot the resuspended lipid on TLC plates. 25 μg triolein was loaded to identify the triglyceride (TG) migration band. Plates are allowed to air dry in a chemical hood for about 20 min. Plates are developed in a solvent mixture (see Recipes section, Kishimoto et al., 2001) in a pre-conditioned TLC chamber. Pre-condition of the chamber involves few additional steps. The chamber is prepared by adding all the components of the solvent mix (Hexane/diethyl ether/acetic acid [70/30/1, v/v]) in the chamber, covering the chamber and mixing vigorously. Additionally, for proper migration of the solvent on the TLC, the inner side of the TLC chamber is lined with two overlapping sheets of Whatman® chromatography paper such that it lines the back and side walls of the chamber (Figure 1). Next, cover the chamber and let it equilibrate for 30 min, before you put the first TLC in the chamber.
Note: Spots should be at least a centimeter above the solvent level.
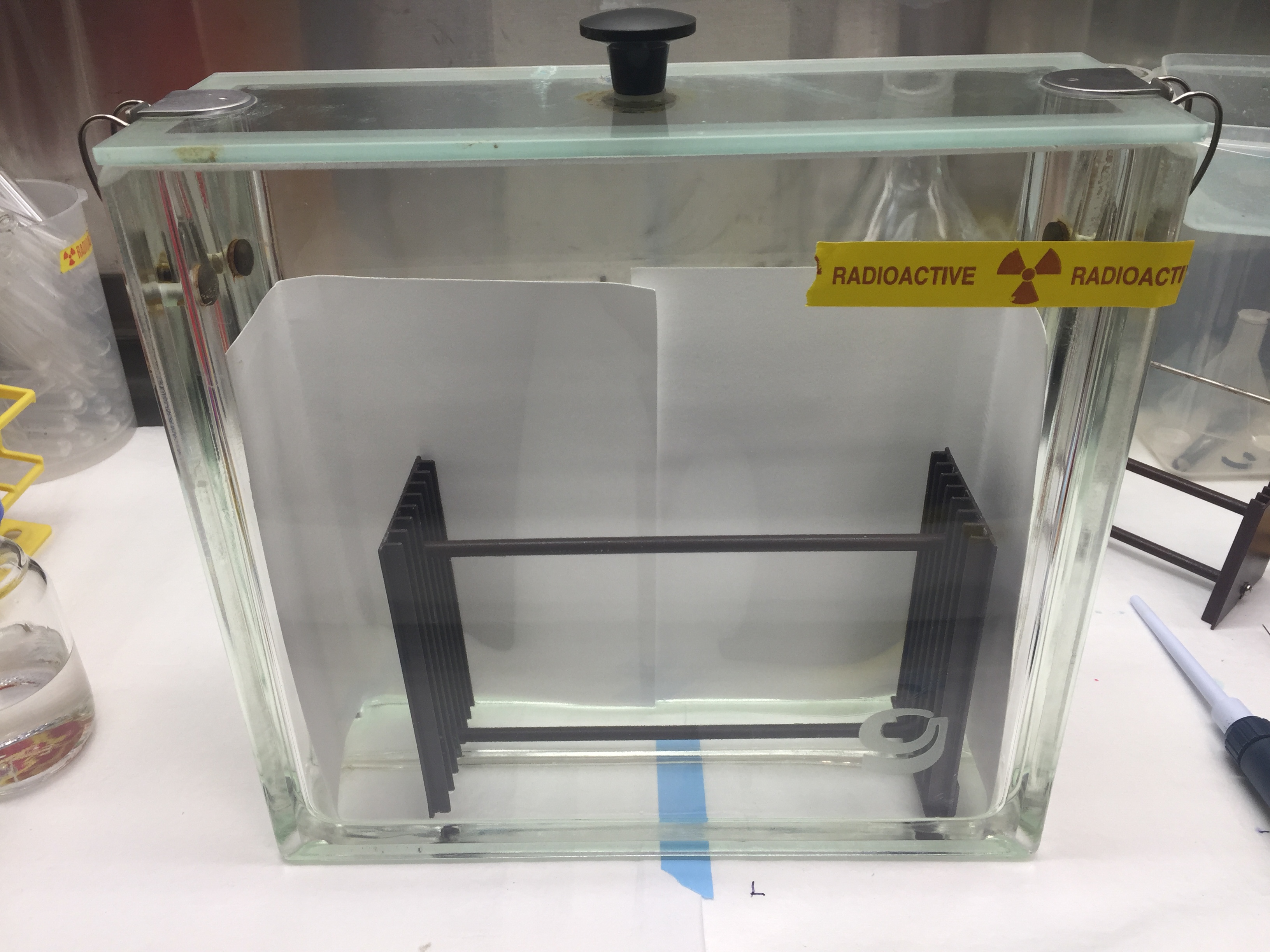
Figure 1. TLC chamber with lid. TLC chamber is lined with two overlapping sheets of Whatman® chromatography paper for saturation of the chamber. The inner Teflon® coated rack allows to simultaneous run several TLC plates.
- Load the dried TLC plate in the chamber and let it run for 10-15 min (depending on the rate of migration of the liquid front). Take the plates out and air dry in a chemical hood.
- For visual identification of TG spots, the TLC plate is sprayed with a solution of cupric sulfate/phosphoric acid (see Recipes section) and allowed to air dry for 30 min. The plates are heated in an oven at 125 °C for 30-45 min (until the TG band is visible). Remove the plate and let it cool. Figure 2 shows the efficiency of TLC to separate different lipid classes.
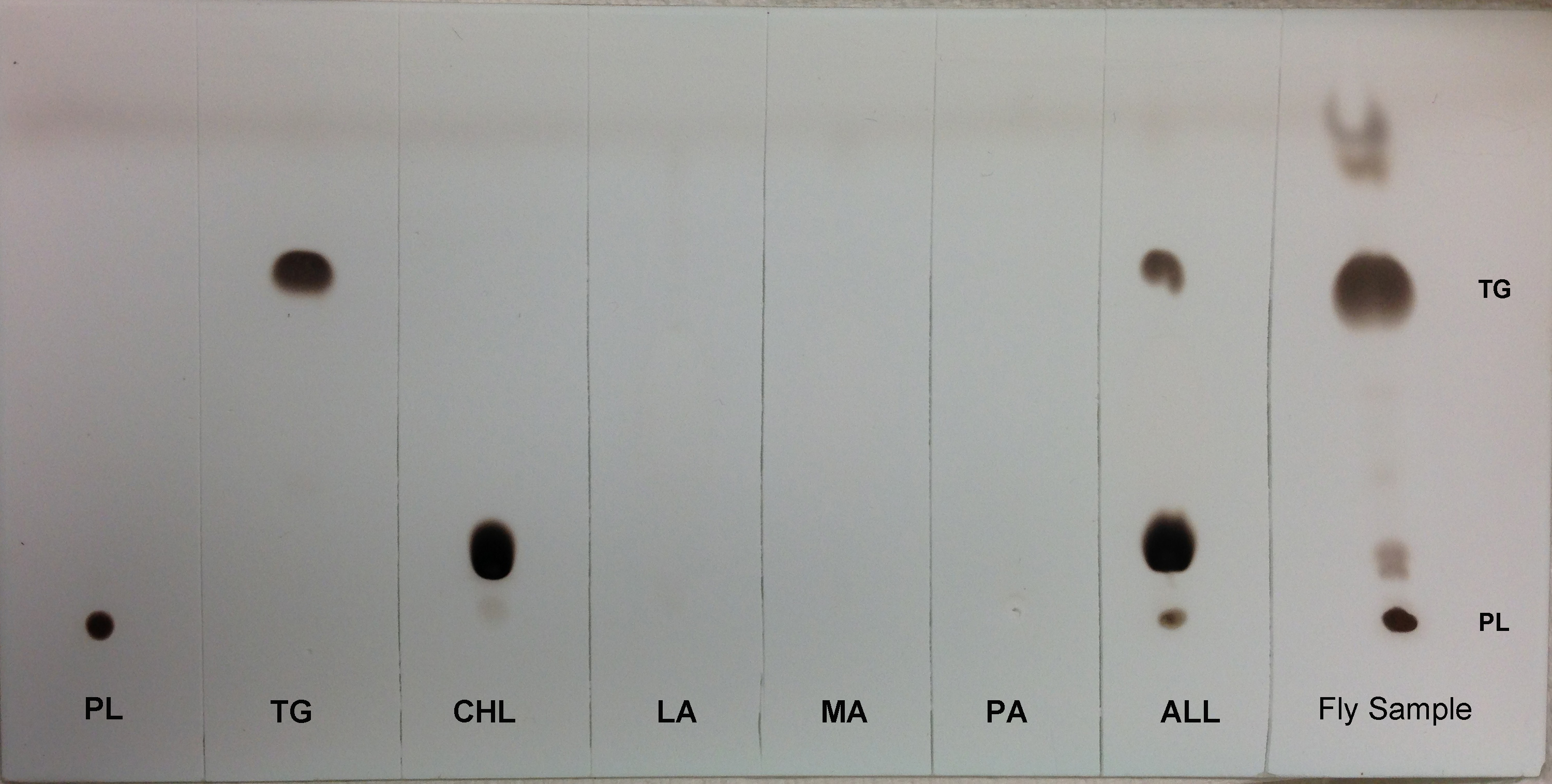
Figure 2. TLC separation of different lipids from adult female Drosophila melanogaster. Total lipid was extracted from 15-20 adult female flies, dried and resuspended in 100 µl of chloroform and loaded on TLC plates. The plates were developed further to visualize migration of different lipids. Lane arrangements are as follows: Lane 1, Phospholipid (PL, a 1:1 mixture of 10 µg phosphatidylcholine and phosphatidylinositol). Lane 2, Triglyceride (TG, 25 µg triolein). Lane 3, Cholesterol (CHL, 10 µg). Lane 4, Lauric acid (LA, 10 µg). Lane 5, Myristic acid (MA, 10 µg). Lane 6, Palmitic acid (PA, 10 µg). Lane 7, ALL (All standards mixed in equal proportions) and Lane 8, 100 µl Fly sample.
- The TG bands from the plates are scraped with a razor blade and transferred to scintillation vials. Add about 0.5 ml of hexane to extract the TG from the silica. Add 3 ml of the scintillation fluid, mix and count the 14C radioactivity. Radioactivity count in the 0 h samples indicate the amount of incorporation of glucose into triglycerides and radioactivity count in 60 h samples indicate the amount of label TG still retained. The difference between 0 h and 60 h samples indicate the breakdown of the labeled triglycerides.
Data analysis
The amount of 14C present in 0 h sample denotes the de novo synthesis of TG and is expressed as CPM/mg fly weight. One experiment containing 5-6 independent samples of 15-25 flies is used to obtain mean ± SEM values. Additional experimental repeats should be carried out to confirm the observed results. Based on the design of the study, one can use Student’s t-test or ANOVA for obtaining the significance. Here, two different diets have been used, and the results (Figure 3) suggest that female flies on low yeast (LY) diet show increased de novo synthesis and a faster breakdown of TG when compared to the flies fed on high yeast (HY) diet.
Note: It’s best to use independent bottles to obtain flies for replicates in one experiment.
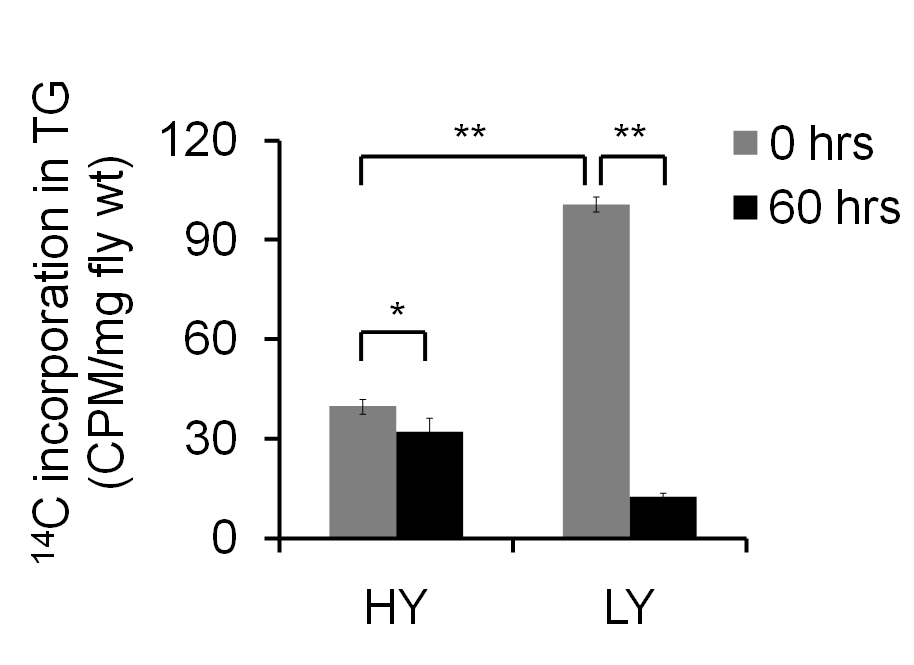
Figure 3. Fat turnover in adult female flies fed on high yeast (HY) and low yeast (LY) diets. The amount of 14C incorporation in 0 h sample indicates the de novo synthesis of TGs. The 60 h sample indicates the breakdown. Student’s t-test was used to measure statistical significance, and error bars denote SEM of five independent preparations (*indicates P < 0.05 and **indicates P < 0.001)
Recipes
- Drosophila food recipes
Detailed fly media recipes can be found in the Supplemental Experimental Procedures section of Katewa et al. (2016).
- Solvent mixture
Hexane/diethyl ether/acetic acid (70/30/1, v/v). Add 105 ml of hexane, 45 ml of diethyl ether and 1.5 ml of acetic acid in the TLC chamber and mix vigorously.
- Cupric sulfate/phosphoric acid solution
8% cupric sulfate in 10% aqueous phosphoric acid
- 0.9% NaCl solution
9 g NaCl in 1 L of double-distilled water
Acknowledgments
SDK acknowledges support from American Federation of Aging Research and Larry L. Hillblom Foundation grants.
References
- Chatterjee, D., Katewa, S. D., Qi, Y., Jackson, S. A., Kapahi, P., and Jasper, H. (2014). Control of metabolic adaptation to fasting by dILP6-induced insulin signaling in Drosophila oenocytes. Proc Natl Acad Sci U S A 111(50): 17959-17964.
- Folch, J., Lees, M. and Sloane Stanley, G. H. (1957). A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem 226(1): 497-509.
- Katewa S.D, Akagi K, Bose N, Rakshit K, Camarella T, Zheng X, Hall D, Davies S, Nelson C, Brem RB, Ramanathan A, Sehgal A, Giebultowicz J.M and Kapahi P (2016). Peripheral circadian clocks mediate dietary restriction dependent changes in lifespan and fat metabolism in Drosophila. Cell Metab. 23(1):143-154.
- Katewa, S. D., Demontis, F., Kolipinski, M., Hubbard, A., Gill, M. S., Perrimon, N., Melov, S. and Kapahi, P. (2012). Intramyocellular fatty-acid metabolism plays a critical role in mediating responses to dietary restriction in Drosophila melanogaster. Cell Metab 16(1): 97-103.
- Kishimoto, K., Urade, R., Ogawa, T. and Moriyama, T. (2001). Nondestructive quantification of neutral lipids by thin-layer chromatography and laser-fluorescent scanning: suitable methods for "lipidome" analysis. Biochem Biophys Res Commun 281(3): 657-662.
Article Information
Copyright
© 2016 The Authors; exclusive licensee Bio-protocol LLC.
How to cite
Katewa, S. D. and Kapahi, P. (2016). Fat Turnover Assay in Drosophila. Bio-protocol 6(21): e1996. DOI: 10.21769/BioProtoc.1996.
Category
Cancer Biology > Cellular energetics > Biochemical assays > Other compound
Biochemistry > Lipid > Lipid measurement
Do you have any questions about this protocol?
Post your question to gather feedback from the community. We will also invite the authors of this article to respond.
Share
Bluesky
X
Copy link